Nearly all living organisms have a collection of bacteria that live within or on their body and provide essential functions, such as aiding digestion and neutralizing toxins. An important community of microbes primarily located in the gastrointestinal tract of animals is known as the gut microbiome. Scientists are working to understand the complex interactions between the gut microbiome and both essential body functions and disease. One way to study the benefits of the microbiome is to analyze organisms with reduced or eliminated microbiomes. These organisms are lacking the bacteria that may help them mediate environmental stressors.
Griffin et al. (2021) presents the development of a methodology to perturb the gut microbiome in bivalves using antibiotics. This methodology can be valuable to further research by providing a technique that produces animals with reduced or eliminated gut microbiomes without killing the hosts. The project began with the PhD work of Dr. Melissa Pierce, a previous student in Prof. Ward’s lab. Her work exposed oysters (Crassostrea virginica) to a cocktail of antibiotics for 4 days, but didn’t observe any significant changes in the diversity of gut microbes. Tyler Griffin’s work extended the exposure period to 3 weeks and used the blue mussel (Mytilus edulis), which is a commonly used bivalve for lab experimentation. As this was his first PhD research project, Griffin reflected that learning when to be independent and learning when to ask for help were some of his biggest challenges. He cited the help of Bridget Holohan and former Ward lab postdoc Dr. Lisa Nigro as invaluable.
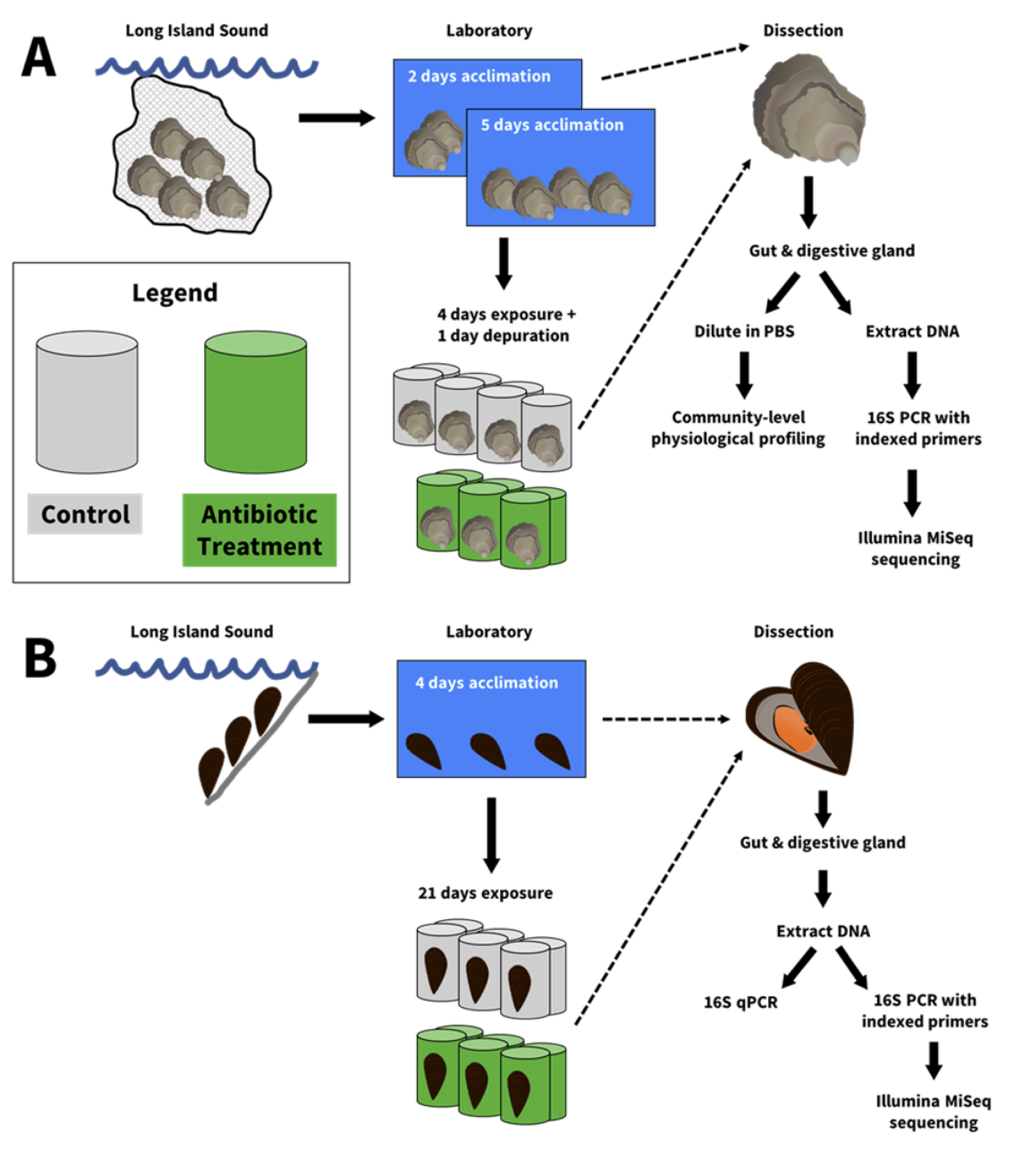
As shown in the flow chart, bivalves were exposed to antibiotics in individual microcosms, or mason jars, and fed a microalgae stock culture that was sterilized by boiling. Three antibiotics were chosen to inhibit a broad group of bacteria. At the end of the experiment, mussels were dissected and analyzed by a few different methods to determine the effect on the microbiome. Through a chance conversation with and help from Dr. Brittany Sprecher, a previous PhD student in Dr. Senjie Lin’s lab, Griffin chose quantitative polymerase chain reaction, or qPCR, which is a technology used for quantifying genes, to check for the total number of bacteria in the gut. qPCR results showed that mussels exposed to antibiotics had a reduced number of bacteria, and other techniques showed reduced microbial species richness and shifts in the whole community composition. Essentially, the antibiotics had successfully reduced the number and type of the bacteria in the gut microbiome, which supported the hypothesis that prolonged exposure to antibiotics can perturb the gut microbiome of bivalves.
Moving forward, Griffin hopes other researchers can use these methods to study other bivalve species, other microbiomes on different body sites, such as the gill, or even other suspension feeders such as gastropods or ascidians.

Griffin, T. W., Pierce, M. L., Nigro, L. M., Holohan, B. A., & Ward, J. E. (2021). An examination of the use of antibiotics as a method to experimentally perturb the microbiota of suspension-feeding bivalves. Invertebrate Biology, 140( 4), e12352. https://doi.org/10.1111/ivb.12352